\def\absarg#1{\left|#1\right|} \def\Earg#1{\mathbf{E}\left[#1\right]} \def\reals{\mathbb{R}} % Real number symbol \def\integers{\mathbb{Z}} % Integer symbol \def\*#1{\mathbf{#1}} \def\m#1{\boldsymbol{#1}} \def\FDR{\widehat{\operatorname{FDR}}} \def\Gsn{\mathcal{N}} \def\Unif{\operatorname{Unif}} \def\Bern{\operatorname{Bern}}

Kris Sankaran
14 | July | 2023
https://github.io/krisrs1128/LSLab
Code: https://go.wisc.edu/s8hdtt
Learning Objectives
We will be able to help collaborators solve scientific problems by adapting modern methods from selective inference.
By the end of this talk, we will be able to:
- Discuss the role of selective inference abstractions in computational genomics settings.
- Interpret p-value histograms and explain how they motivate the Benjamini-Hochberg procedure.
- Improve power in large-scale inference by focusing on the most promising contexts.
- Diagnose miscalibration in large-scale inference and apply data splitting to address it.
Large Scale Inference
Notation
- Hypotheses of interest: H_{1}, \dots, H_{M}. Some of them are non-null, but you don't know which.
- Associated p-values: p_{1}, \dots, p_{M}.
Goal: Reject as many non-null hypotheses as possible while controlling the False Discovery Rate,
\begin{align*} \text{FDR} := \Earg{\frac{\absarg{\text{False Positives}}}{\absarg{\text{Rejections}}\vee 1}} \end{align*}
Examples
- Microbiome: Is taxon m associated with the development of autism in infants?
- Epigenetics: Is CpG site m differentially methlyated among smokers?
- Cancer: Is elevated immune cell expression of gene m associated with improved survival rates?
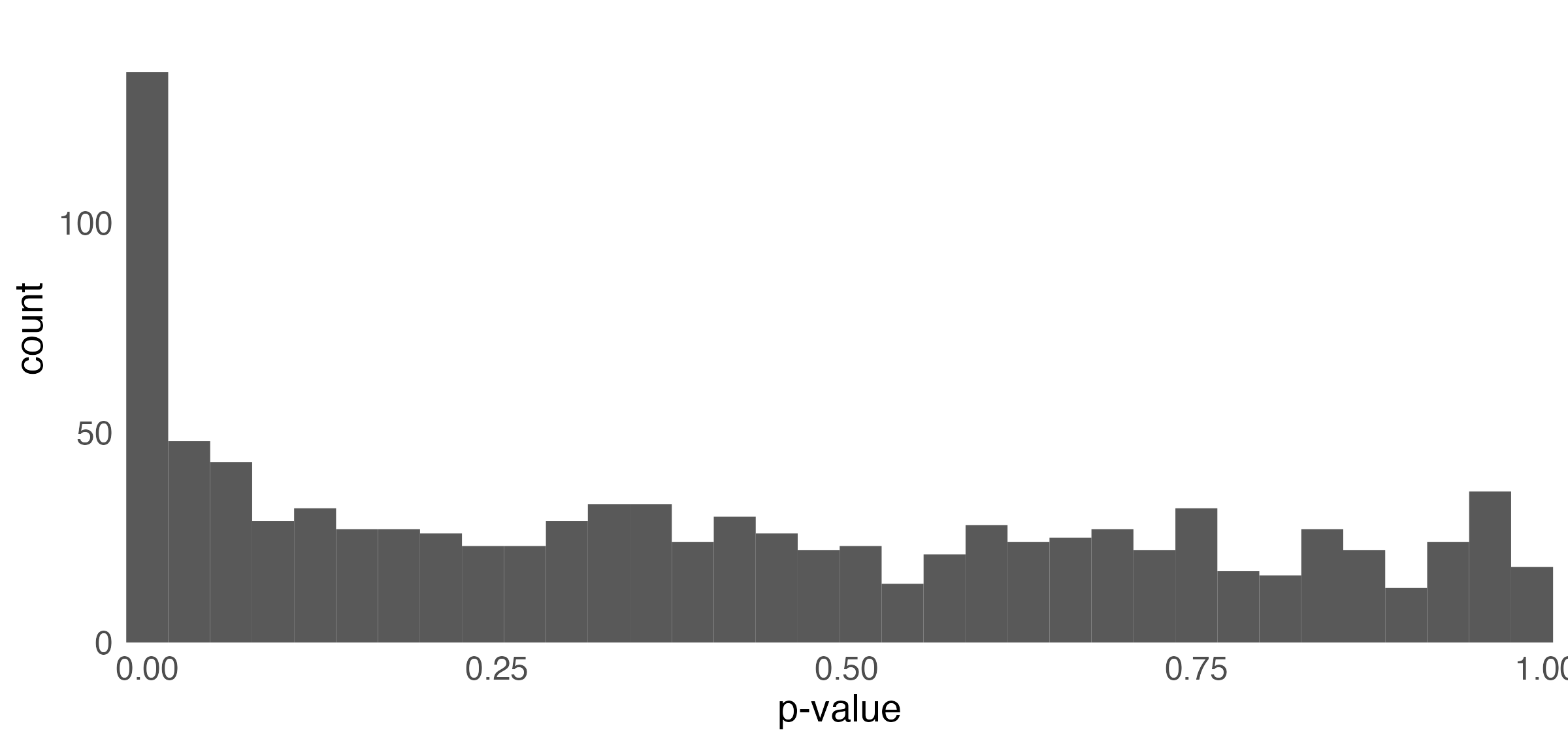
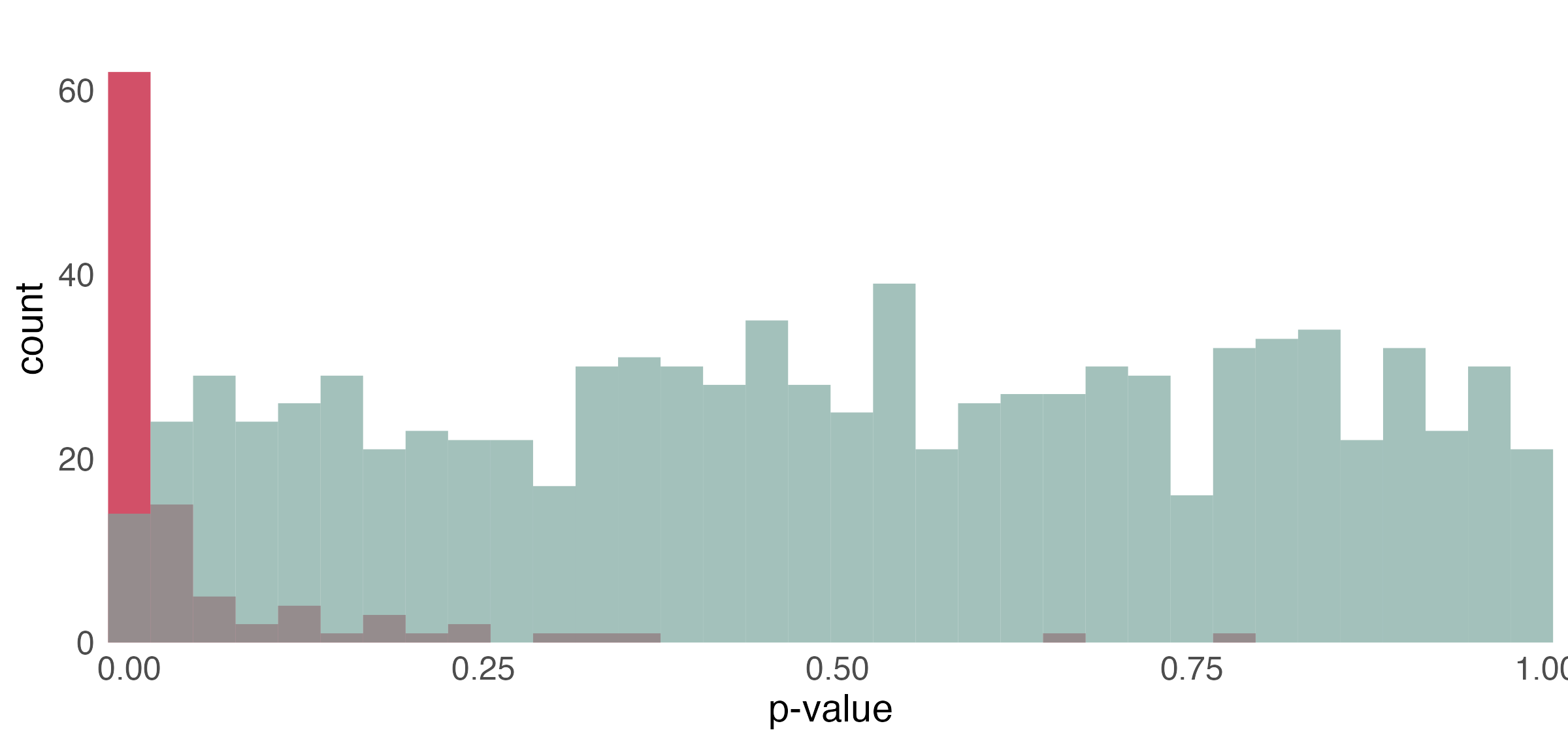
p-value histogram
- Under the null, the p-values follow a uniform distribution.
- The spike near 0 \implies true alternative hypotheses
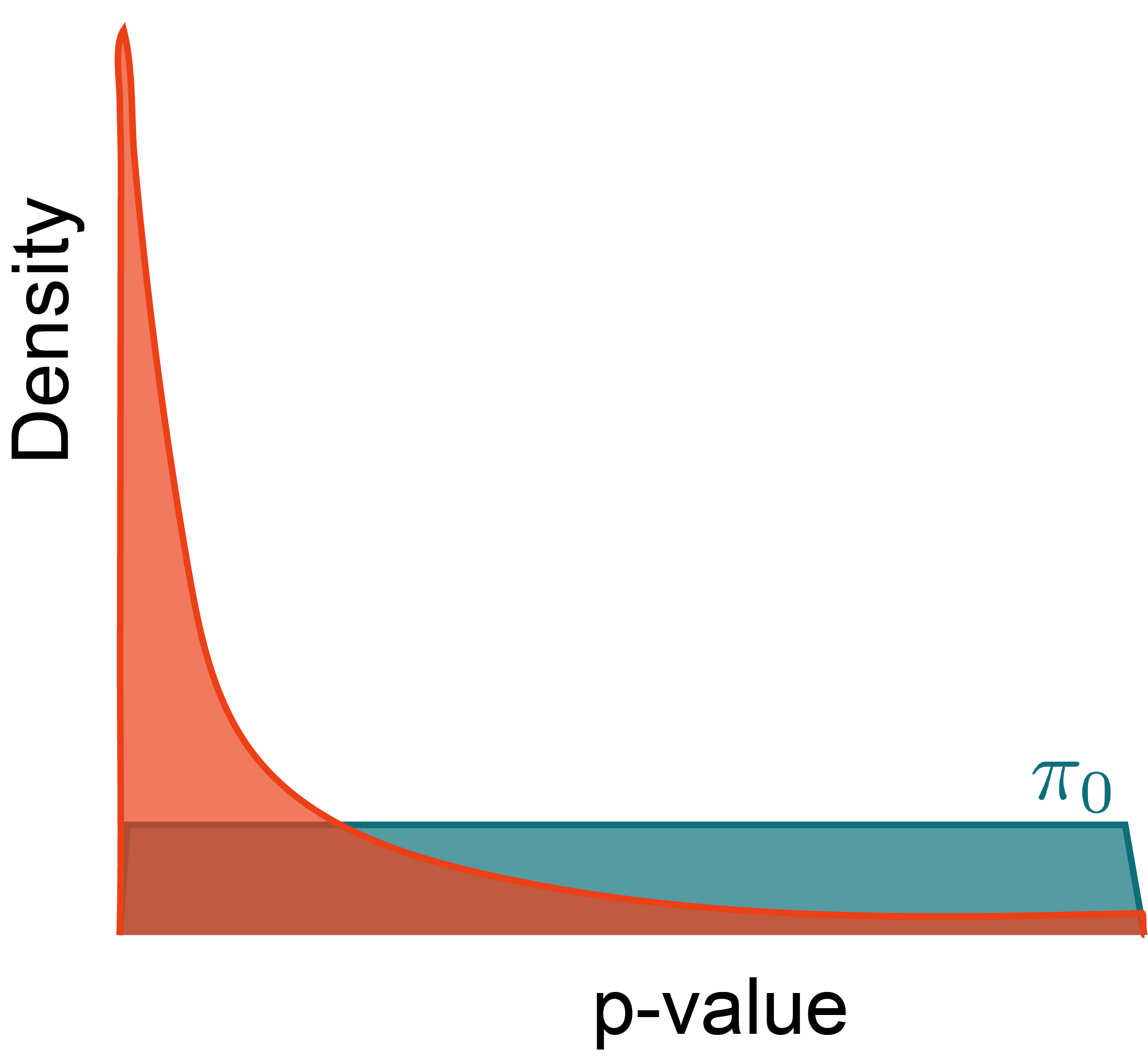
- \pi_{0} refers to the true proportion of nulls
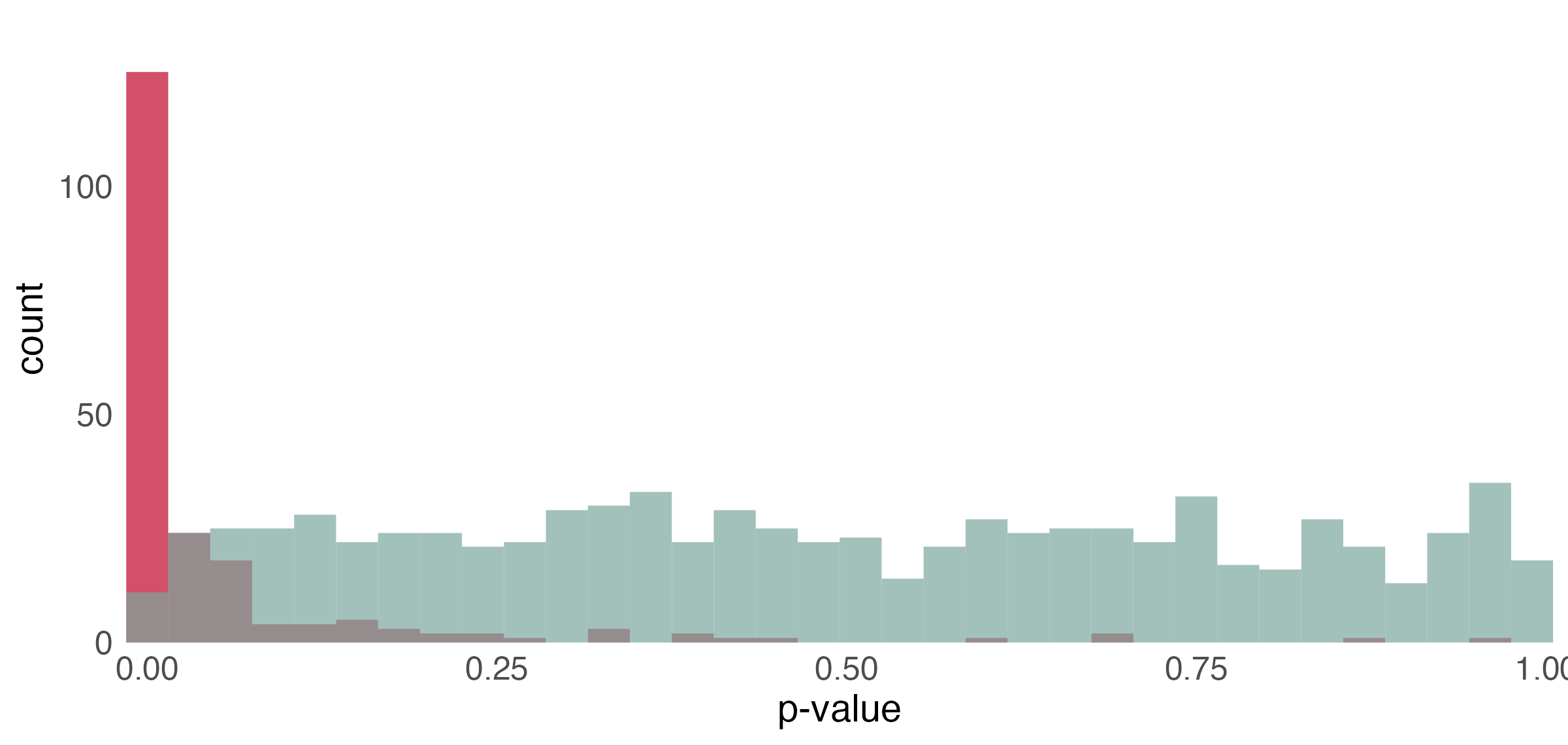
p-value histogram
- Under the null, the p-values follow a uniform distribution.
- The spike near 0 \implies true alternative hypotheses
- \pi_{0} refers to the true proportion of nulls
p-value histogram
- Under the null, the p-values follow a uniform distribution.
- The spike near 0 \implies true alternative hypotheses
- \pi_{0} refers to the true proportion of nulls
Benjamini-Hochberg
The Benjamini-Hochberg (BH) procedure [1] controls the FDR at level q.
- Sort the p-values: p_{(1)} \leq p_{(2)} \leq \dots \leq p_{(M)}
- Find the largest i such that p_{(i)} \leq \frac{i q}{M}
- Reject hypotheses associated with p_{(1)} \leq \dots \leq p_{(i)}
Why?
At any threshold t, estimate the FDR using relative areas from the null and alternative densities.
Why?
At any threshold t, estimate the FDR using relative areas from the null and alternative densities.
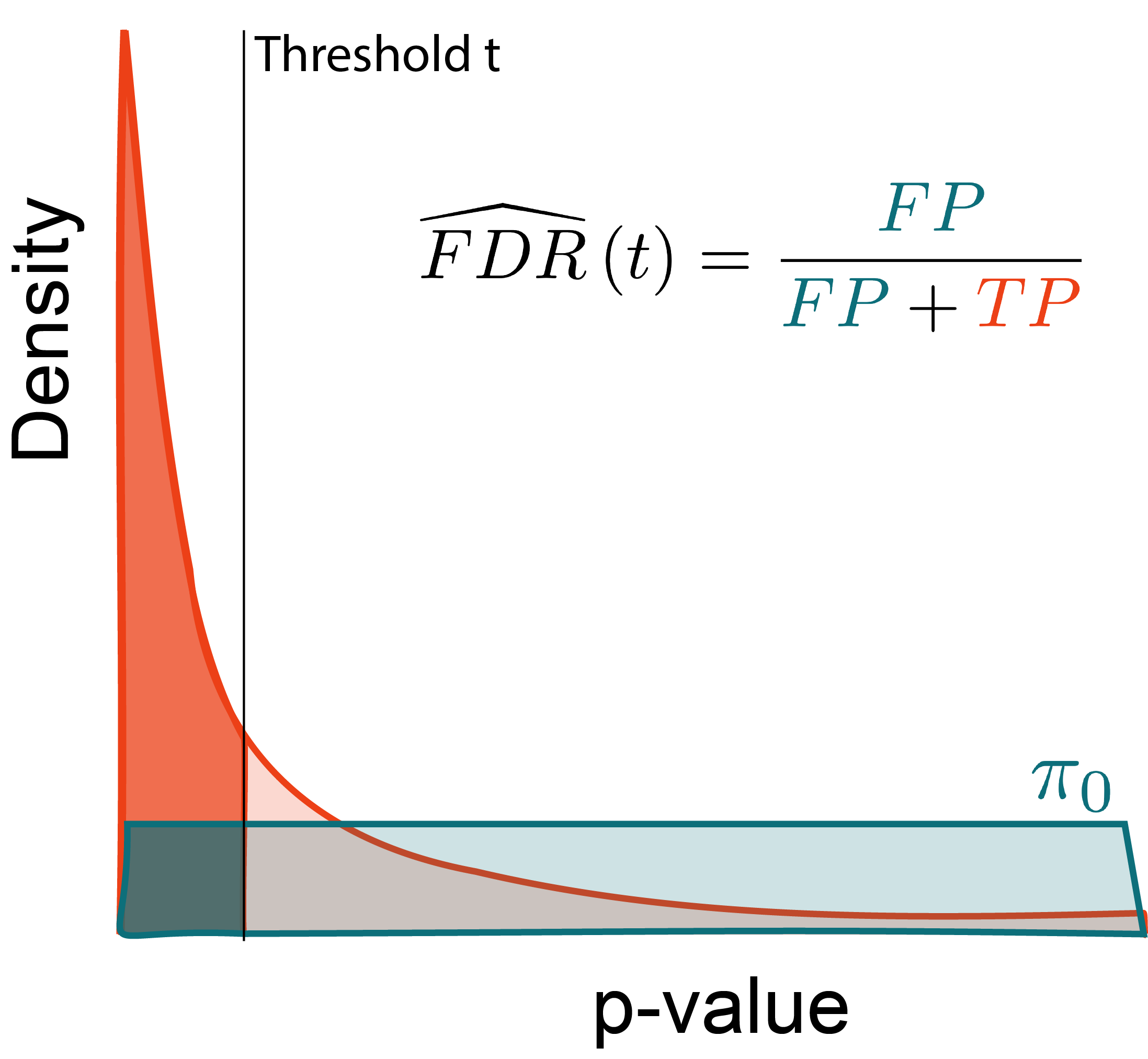
Why?
Let R\left(t\right) be the number of rejected hypotheses at threshold t. Then,
\begin{align*} \FDR\left(t\right) &= \frac{\pi_{0}Mt}{R\left(t\right)} \end{align*}

Why?
Maximize the number of rejections while limiting false discoveries.
Optimize: \begin{align*} \text{maximize } &R\left(t\right) \\ \text{subject to } &\FDR\left(t\right) \leq q \end{align*}

Why?
Maximize the number of rejections while limiting false discoveries.
Optimize: \begin{align*} \text{maximize } &t \\ \text{subject to } &\FDR\left(t\right) \leq q \end{align*}

Why?
Larger thresholds let more hypotheses through.
Optimize: \begin{align*} \text{maximize } & t \\ \text{subject to } &\FDR\left(t\right) \leq q \end{align*}
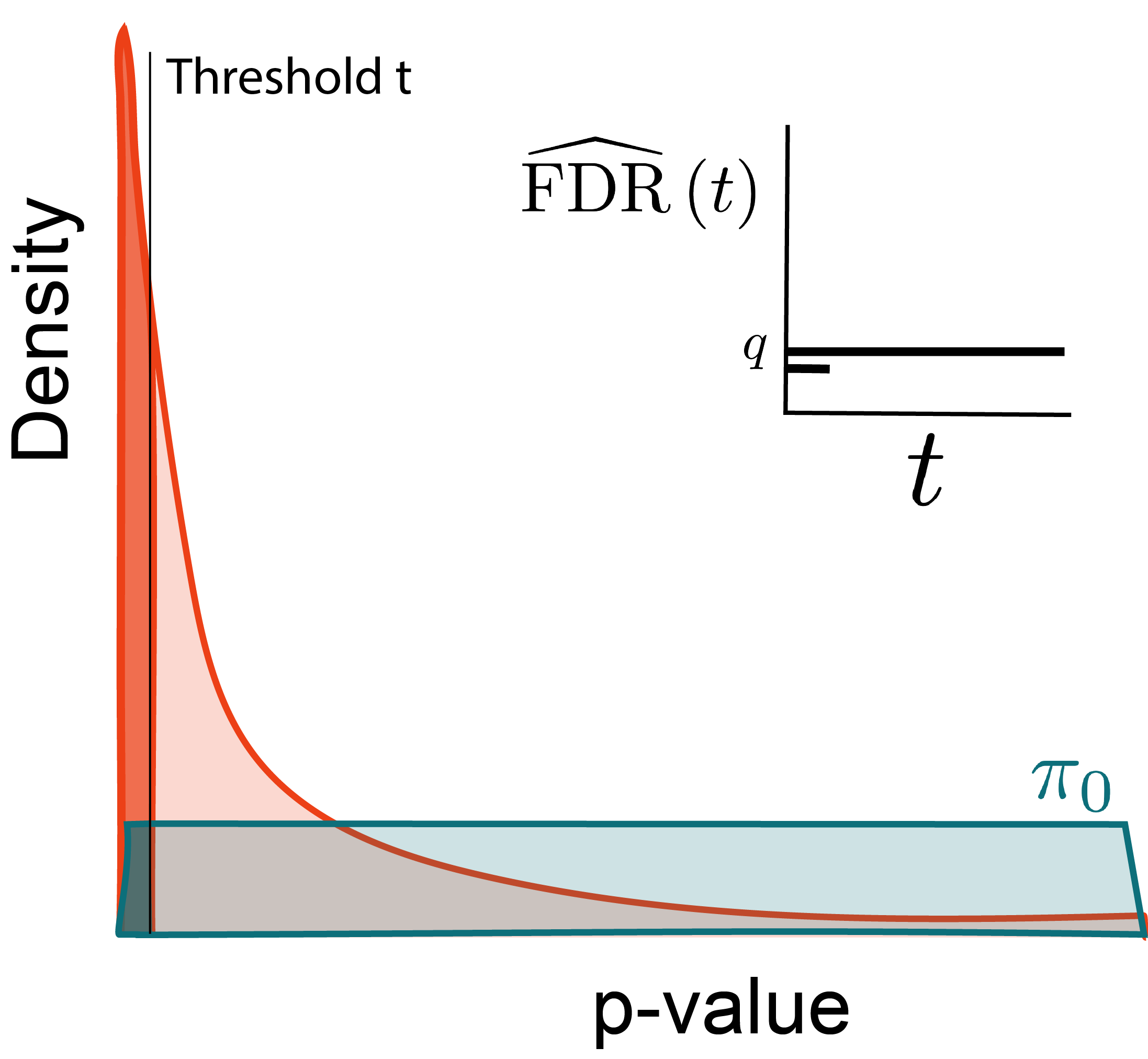
Why?
Larger thresholds let more hypotheses through.
Optimize: \begin{align*} \text{maximize } & t \\ \text{subject to } &\FDR\left(t\right) \leq q \end{align*}
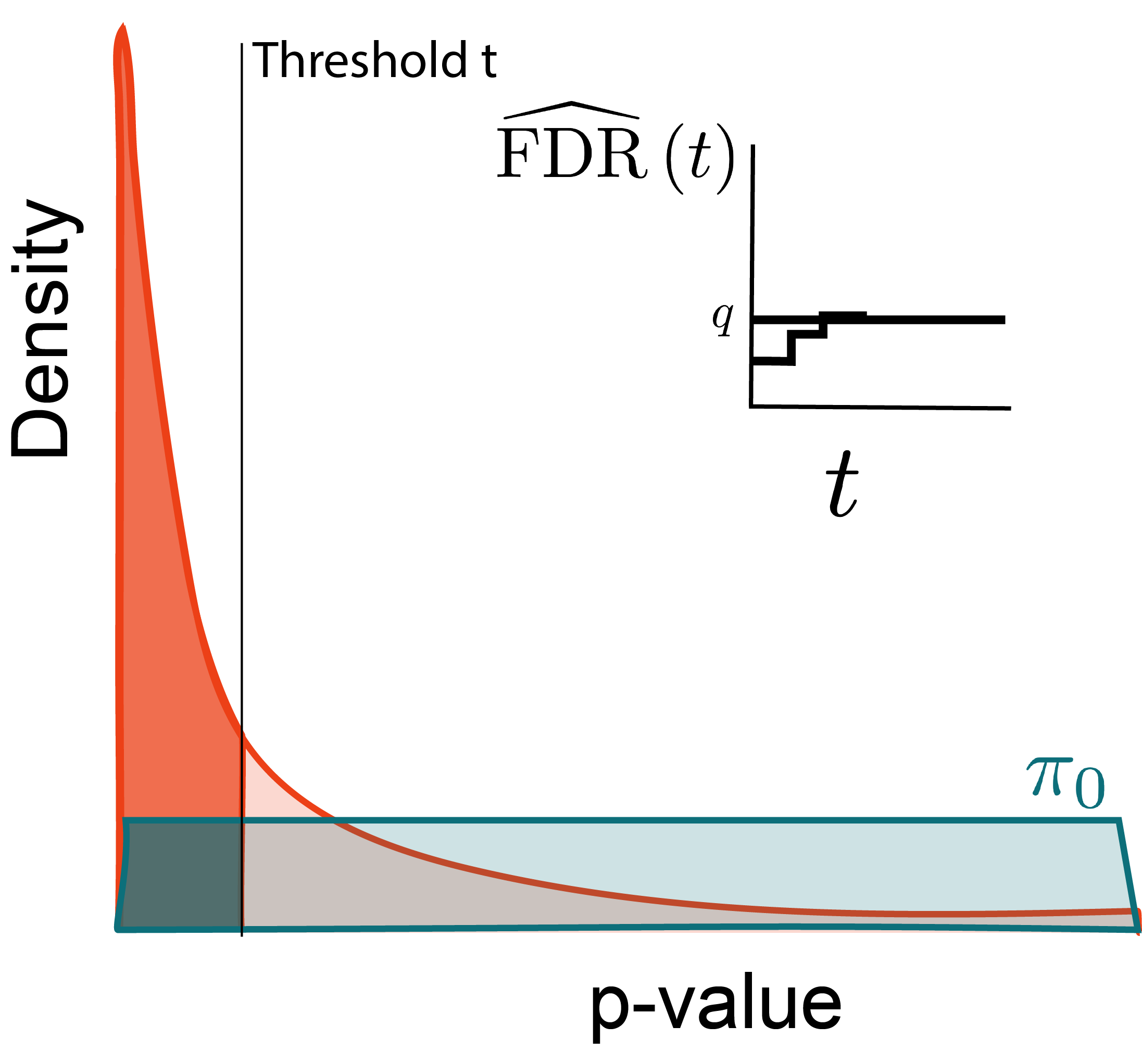
Why?
Larger thresholds let more hypotheses through.
Optimize: \begin{align*} \text{maximize } &t \\ \text{subject to } &\FDR\left(t\right) \leq q \end{align*}
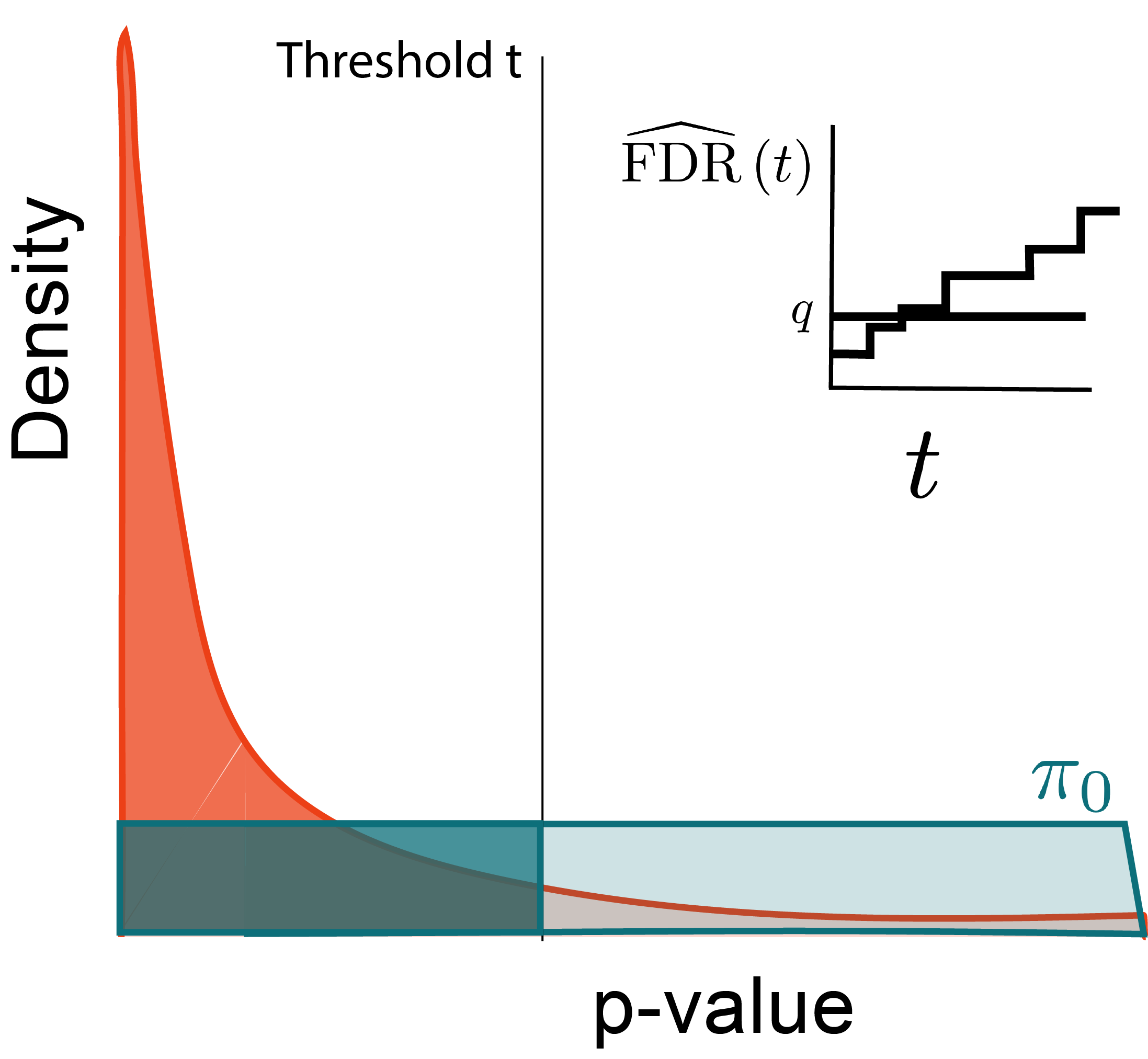
Why?
Look for the largest p-value satisfying this inequality.
Optimize: \begin{align*} \text{maximize } &t \\ \text{subject to } &\FDR\left(t\right) \leq q \end{align*}
If we substitute t = p_{(i)}, then
\begin{align*} &\frac{\pi_{0}M p_{(i)}}{i} \leq q \\ \implies & p_{(i)} \leq \frac{i q}{\pi_{0} M} \\ \implies & p_{(i)} \leq \frac{i q}{M} \end{align*}
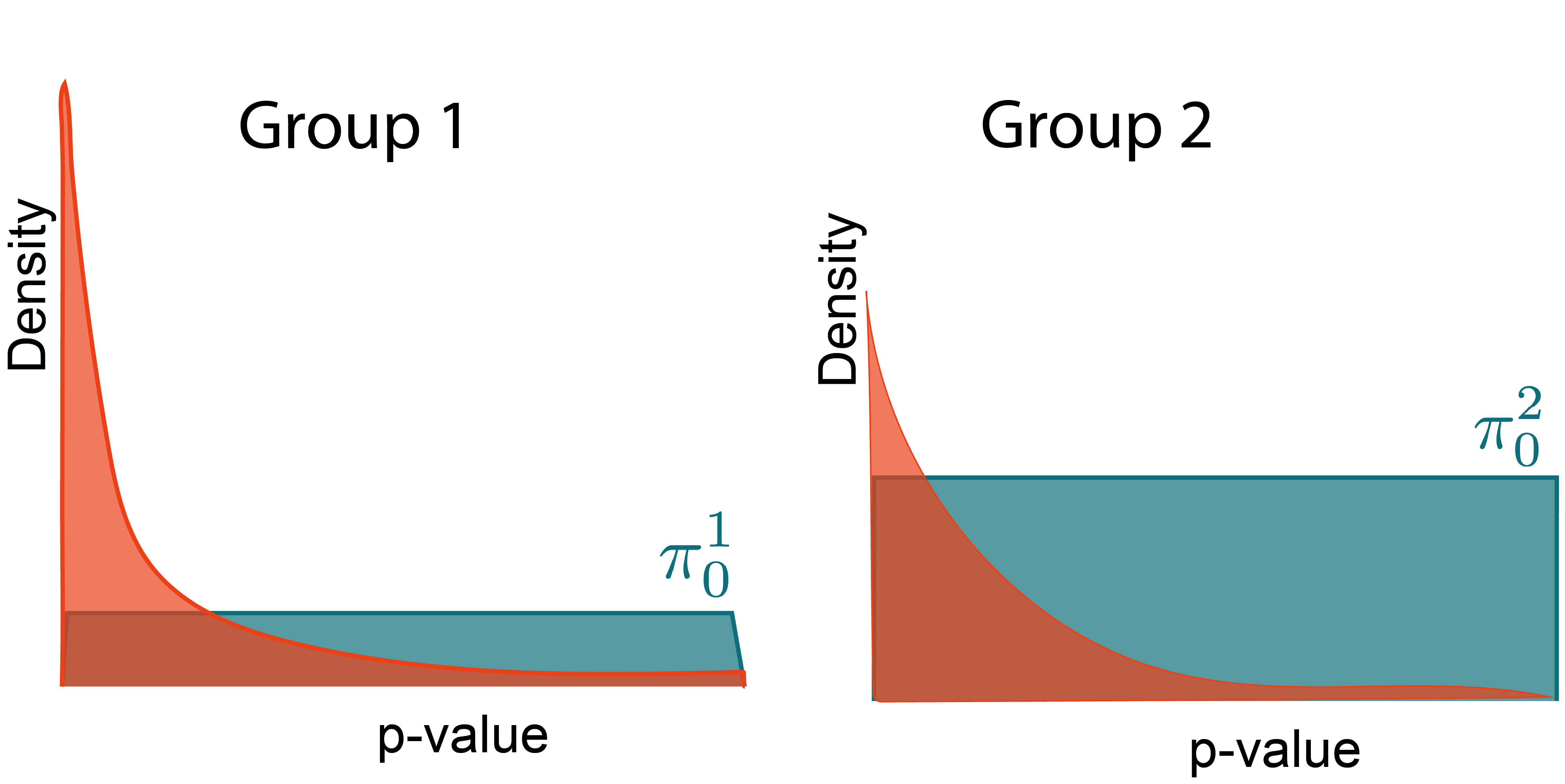
Power through Context
Hypotheses are not identical
- We may know in advance that some hypotheses are more likely to be rejected than others.
- Prior literature
- Number of mapped reads
- This information can improve power. We'll discuss [2], but see [3; 4; 5; 6]
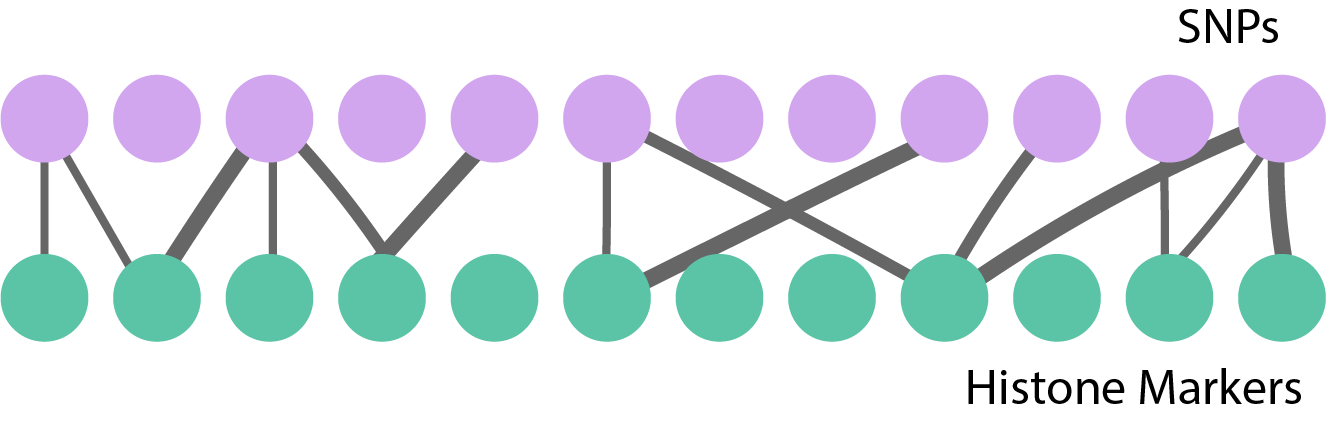
Example: Testing Pairs
Suppose we are testing associations between,
- SNPs x_{1}, \dots, x_{I}
- Histone Markers y_{1}, \dots, y_{J}
H_{m}: \text{Cor}\left(x_{i}, y_{j}\right) = 0.
Even for moderate I, J, this is many tests!
Grouping by Distance
- Group the distances into bins 1, \dots, G.
- We use different thresholds \*t = \left(t_{1}, \dots, t_{G}\right) across groups.
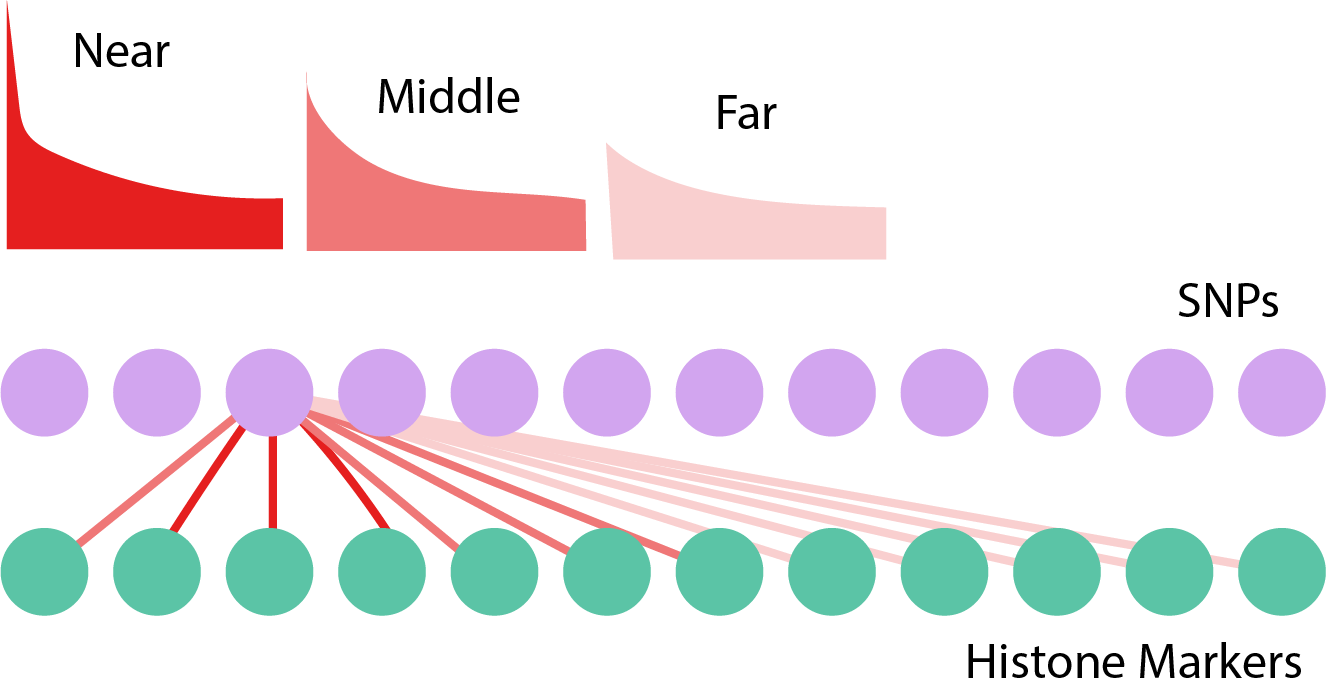
Grouping by Distance
- Group the distances into bins 1 , \dots, G.
- We use different thresholds \*t = \left(t_{1}, \dots, t_{G}\right) across groups.
\begin{align*} R\left(\*t\right) &:= \sum_{g} R_{g}\left(t_{g}\right) \\ \FDR\left(\mathbf{t}\right) &:= \frac{\sum_{g = 1}^{G} m_{g}t_{g}}{R\left(\*t\right)} \end{align*}
\begin{align*} \text{maximize } &R\left(\*t\right) \\ \text{subject to } &\FDR\left(\*t\right) \leq q \end{align*}
Code Demo
We simulated a SNP-histone marker association dataset x_{n} \in \{0, 1\}^{I} and y_{n} \in \reals^{J}.
\begin{align*} y_{n} &= B x_{n} + \epsilon_{n} \\ \epsilon_{n} &\sim \mathcal{N}\left(0, 0.1^2\right) \\ x_{ni} &\sim \Bern\left(\frac{1}{2}\right) \\ B_{nj} &\sim \Bern\left(0.4 \exp\left(- \frac{1}{10}\left|\frac{i}{I} - \frac{j}{J}\right|\right)\right) \\ \end{align*}
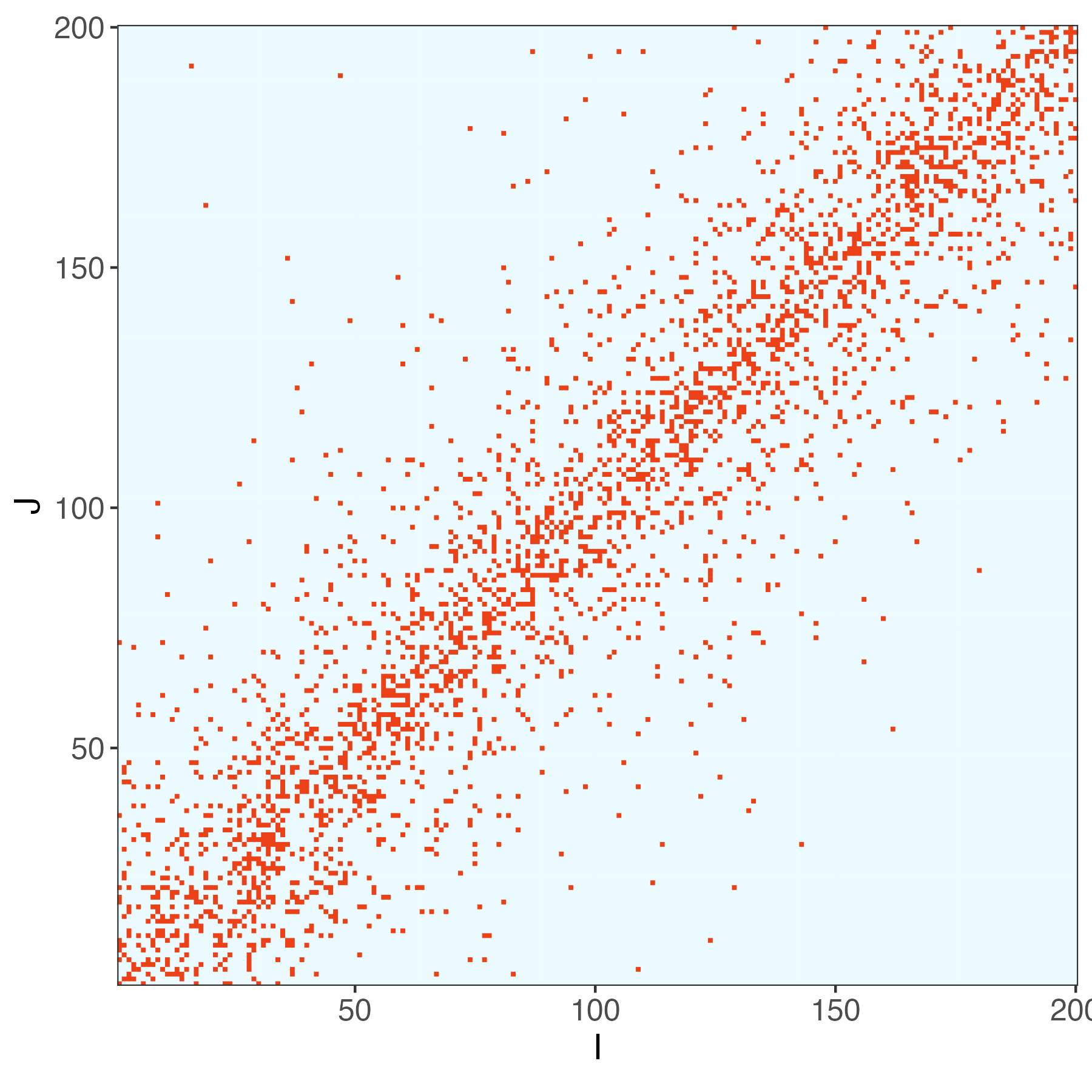
Code Demo
We simulated a SNP-histone marker association dataset x_{n} \in \{0, 1\}^{I} and y_{n} \in \reals^{J}.
\begin{align*} y_{n} &= B x_{n} + \epsilon_{n} \\ \epsilon_{n} &\sim \mathcal{N}\left(0, 0.1^2\right) \\ x_{ni} &\sim \Bern\left(\frac{1}{2}\right) \\ B_{nj} &\sim \Bern\left(0.4 \exp\left(- \frac{1}{10}\left|\frac{i}{I} - \frac{j}{J}\right|\right)\right) \\ \end{align*}
Code Demo
We can estimate \FDR\left(\*t\right) with a few lines of code:
m <- map_dbl(p_values, length)R <- map2_dbl(p_values, thresholds, ~ sum(.x < .y))sum(m * thresholds) / Rp_valuesis a list of p-values for each group gthresholdsis a vector storing \*t
Code Demo
We can have more generous thresholds for p-values between nearby pairs. This invests the most "budget" on the promising contexts.
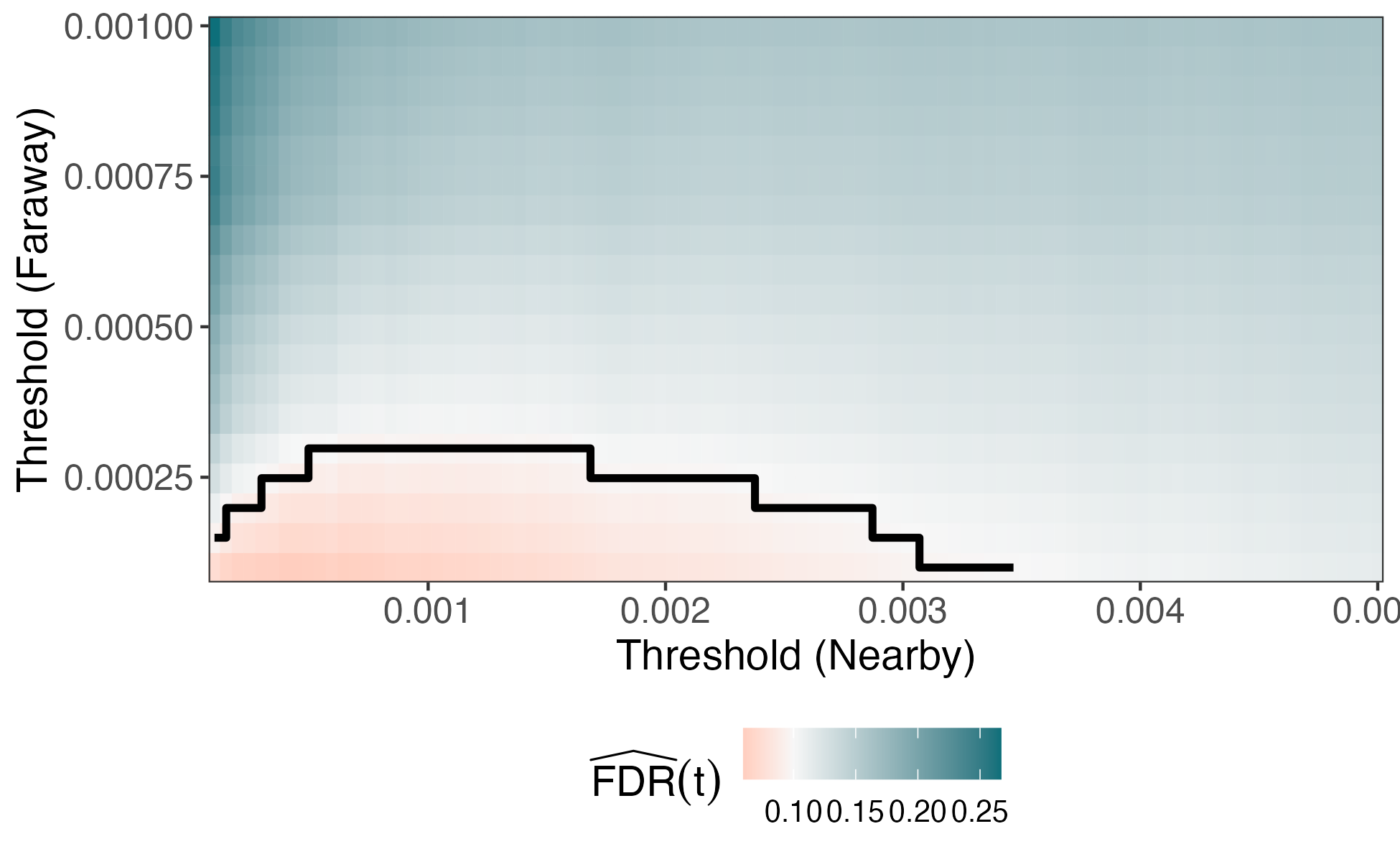
Code Demo
We can have more generous thresholds for p-values between nearby pairs. This invests the most "budget" on the promising contexts.
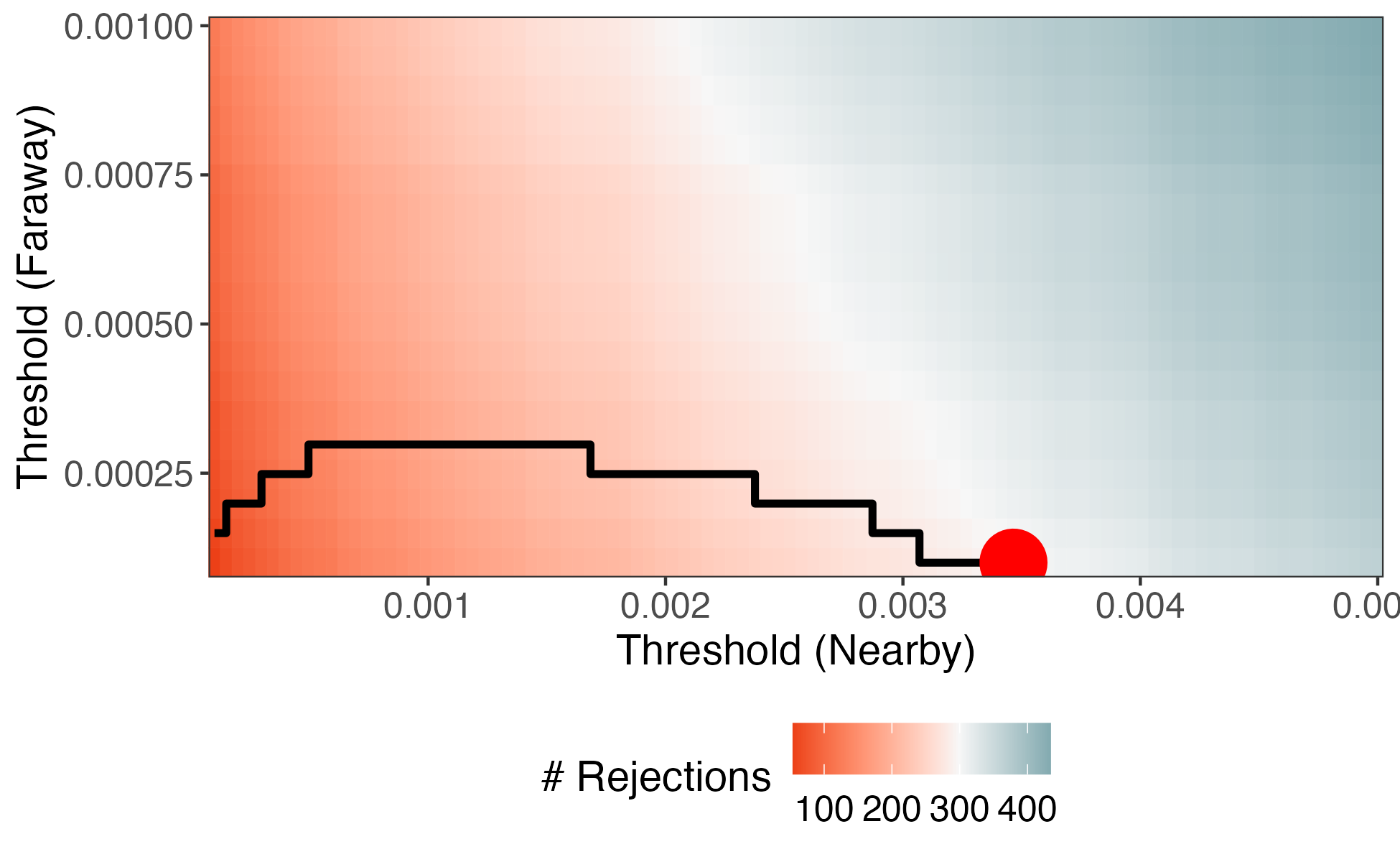
Optimization
- In practice, we would optimize \*t, rather than scanning the entire space.
- The number of rejections can be derived from the CDF of the p-value histogram, and the problem can be formulated as a convex optimization.
- We haven't discussed it, but it is important to cross-weight to ensure independence between the thresholds and p-values.
Calibration through Splitting
Example
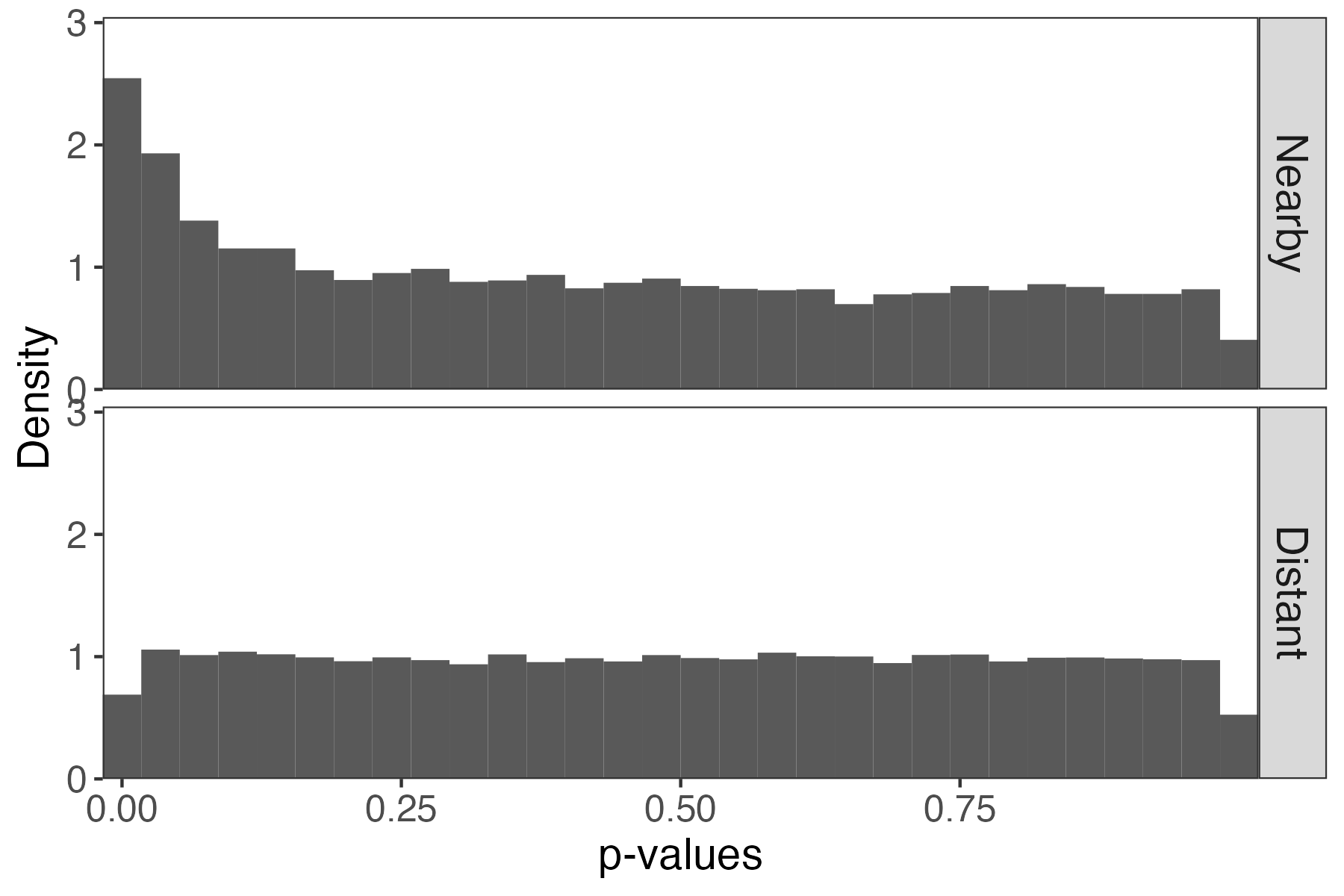
Imagine gene m in sample n is distributed according to, \begin{align*} x_{nm} \vert \text{disease t} \sim \text{NB}\left(\mu_{mt}, \varphi\right) \end{align*} so that expression potentially varies according to disease status.
\begin{align*} H_{0m}: \mu_{m0} = \mu_{m1} \\ H_{1m}: \mu_{m0} \neq \mu_{m1} \end{align*}
Example
- We used the
edgeRpackage to normalize these data (TMM + log transform) [7], inspired by the simulation in [8] - Notice the nonuniformity of the null p-values from the two-sample t-tests
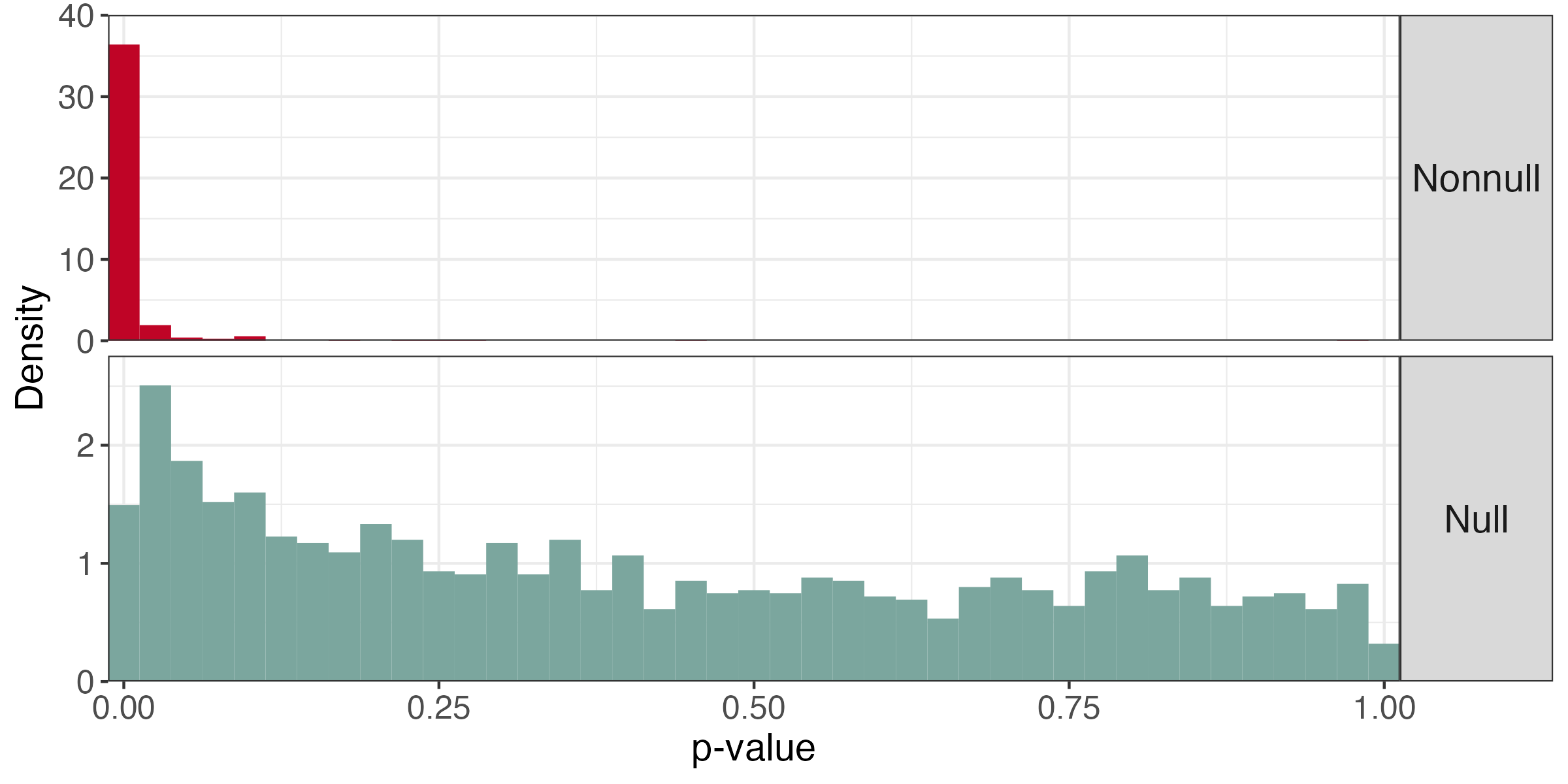
Key Idea
We can avoid this problem if we bypass p-values altogether.
- Significance can be deduced from test statistics directly
- We do not have to use BH to ensure FDR control
This idea is highly adaptable. We'll discuss [9], but see also [10; 11].
Testing \to Regression
We work with the original data \*X \in \reals^{N \times M} and \*y \in \reals^{N}. For example,
- y_{n}: Disease state for patient n
- x_{nm}: Expression level for gene m.
Consider regressing \*y = \*X \*\beta + \epsilon. The null hypothesis is that \beta_{m} = 0.
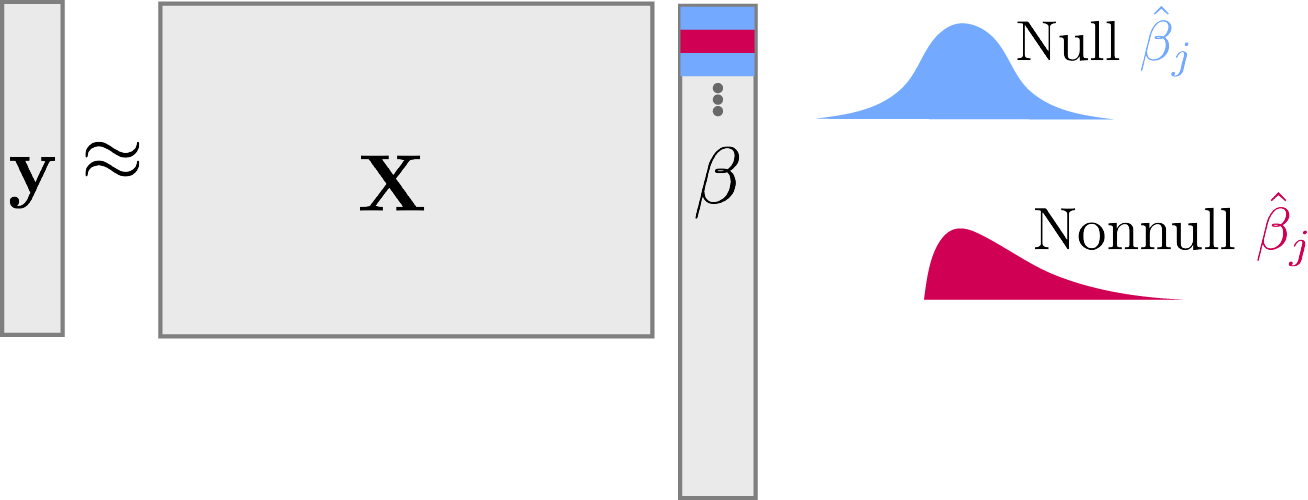
Symmetry and Mirrors
Randomly split rows into \left(\*X^{(1)}, \*y^{(1)}\right) and \left(\*X^{(2)}, \*y^{(2)}\right).
Mirror statistics have the form, \begin{align*} T_{m} &= \text{sign}\left(\hat{\beta}^{(1)}_{m}\hat{\beta}_{m}^{(2)}\right)f\left(\absarg{\hat{\beta}^{(1)}_{m}}, \absarg{\hat{\beta}^{(2)}_{m}}\right) \end{align*}
where \hat{\beta}^{(j)} are estimated on the separate splits.
Symmetry and Mirrors
Mirror statistics measure the agreement in estimates across splits.
Example: \text{sign}\left(\hat{\beta}^{(1)}_{m}\hat{\beta}_{m}^{(2)}\right)\left[\absarg{\hat{\beta}_{m}^{(1)}} + \absarg{\hat{\beta}_{m}^{(2)}}\right]
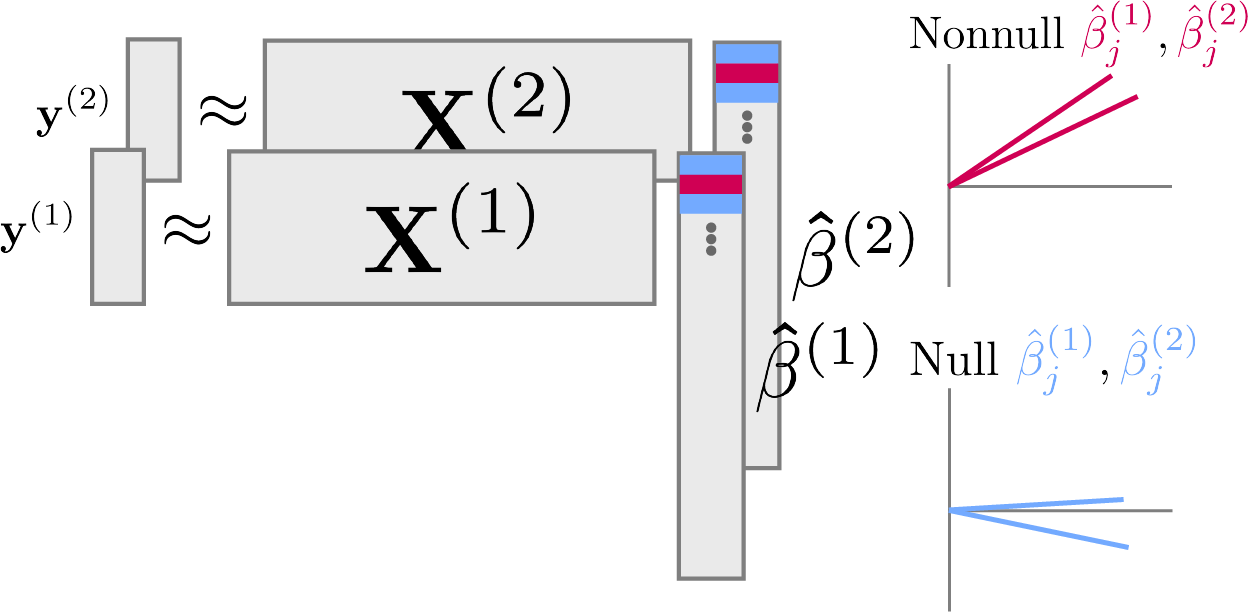
Symmetry and Mirrors
Mirror statistics measure the agreement in estimates across splits.
Example: \text{sign}\left(\hat{\beta}^{(1)}_{m}\hat{\beta}_{m}^{(2)}\right)\left[\absarg{\hat{\beta}_{m}^{(1)}} + \absarg{\hat{\beta}_{m}^{(2)}}\right]
ix <- sample(nrow(x), 0.5 * nrow(x))beta1 <- train_fun(x[ix], y[ix])beta2 <- train_fun(x[-ix], y[-ix])sign(beta1 * beta2) * (abs(beta1) + abs(beta2))Mirror Histogram
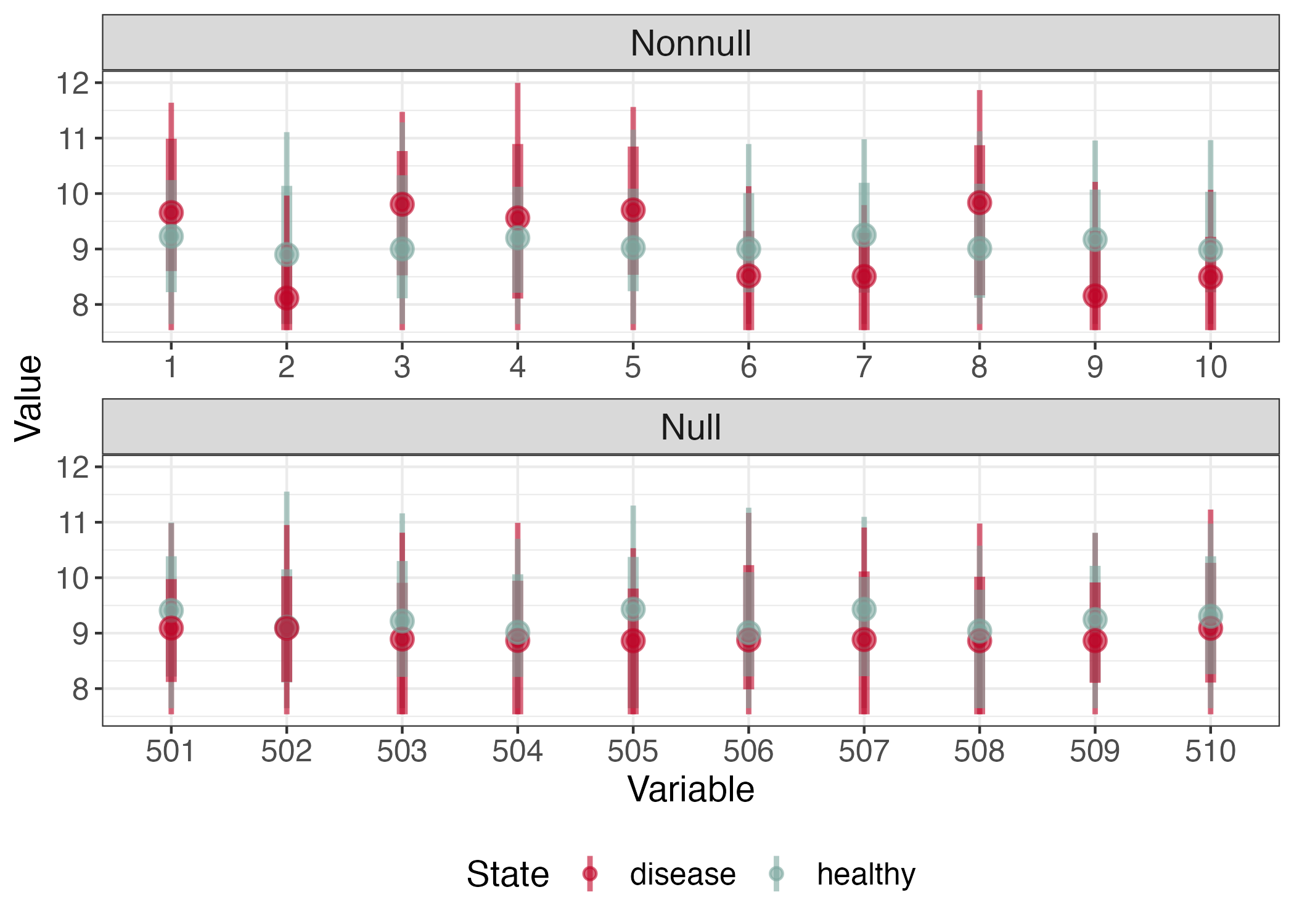
Let's revisit the negative binomial problem. Which hypotheses should we reject?
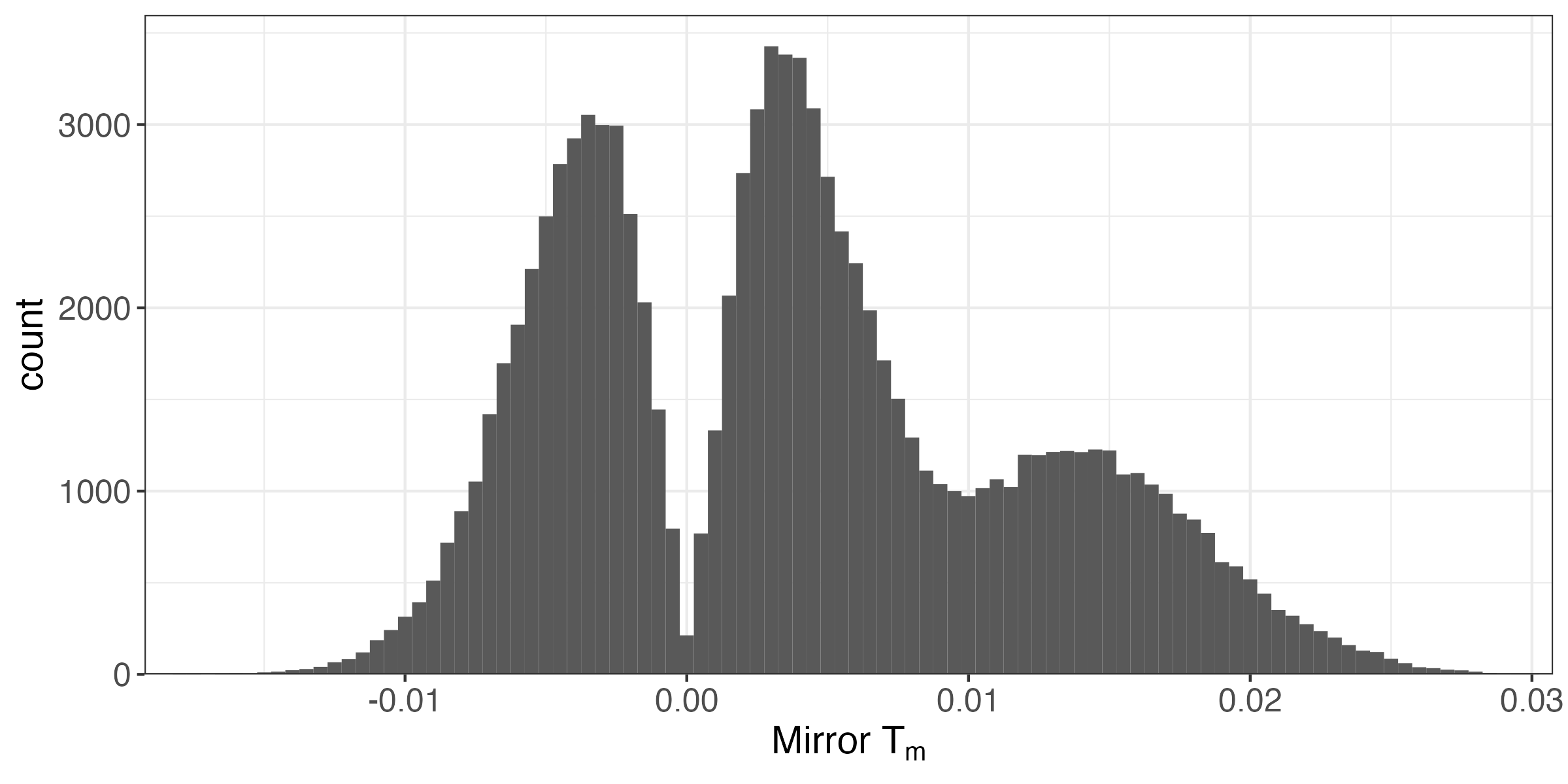
Mirror Histogram
A reasonable strategy is to reject for all T_{m} in the far right tail. How should we pick the threshold?
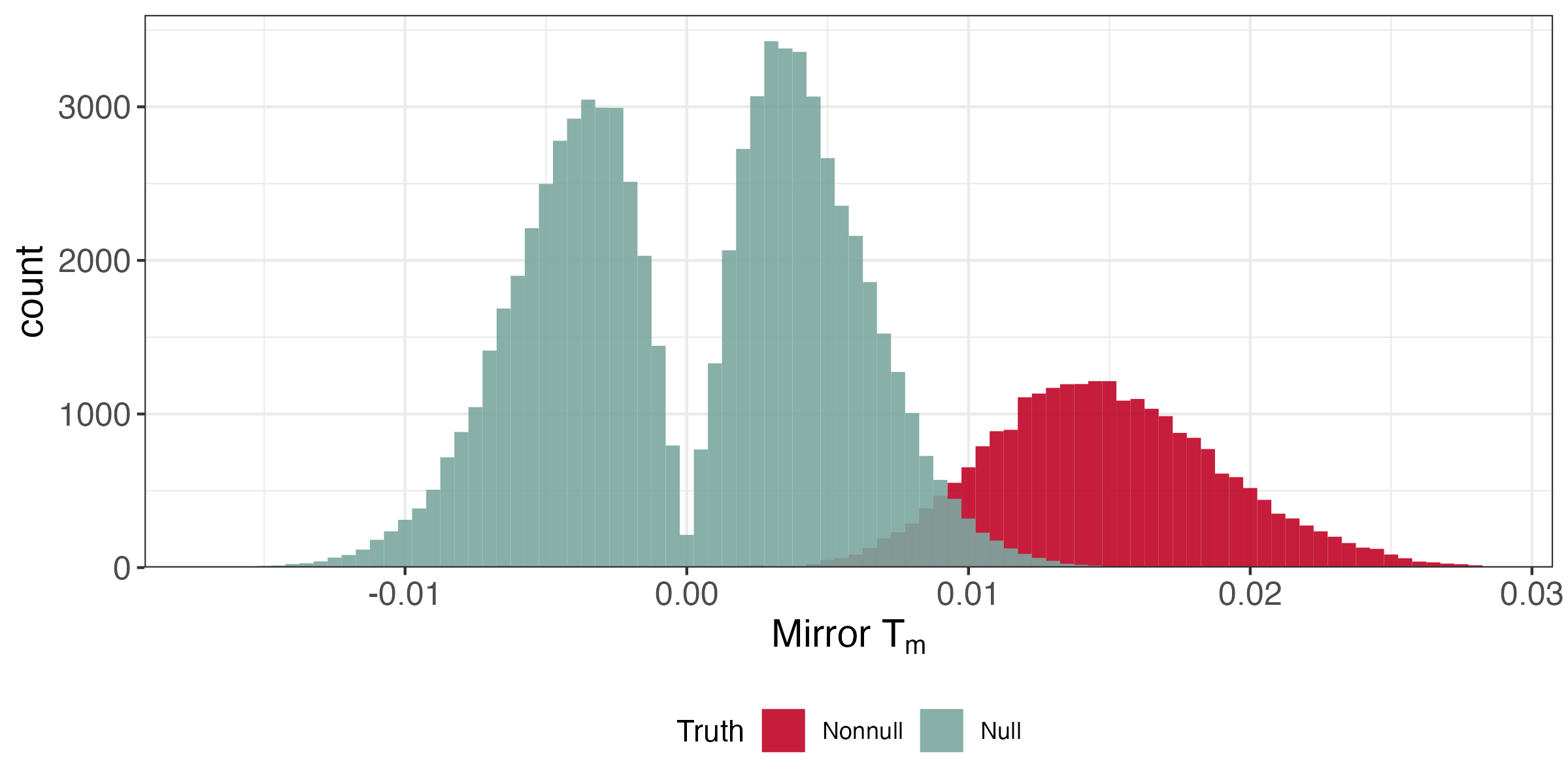
FDR Estimation
By symmetry, the size of the left tail \approx the number of nulls in the right tail,
\begin{align*} \FDR\left(t\right) &= \frac{\absarg{T_{m} < -t}}{\absarg{T_{m} > t}} \end{align*}
which suggests solving, \begin{align*} \text{maximize } &t \\ \text{subject to } &\FDR\left(t\right) \leq q \end{align*}

Multiple Splits
Issue:
- A downside is that sample splitting samples reduces power.
- Power can be recovered by aggregating over multiple iterations of splitting.
Main Idea:
- Create K different pairs of random splits
- Let \hat{S}_{k} be the selected features when using mirror statistics on the k^{th} random split
- Look for consensus across the \hat{S}_{k}
- (See backup slides for details)
Code Demo
In our Negative Binomial example, this leads to FDR control and reasonable power. Here are the results over multiple runs.
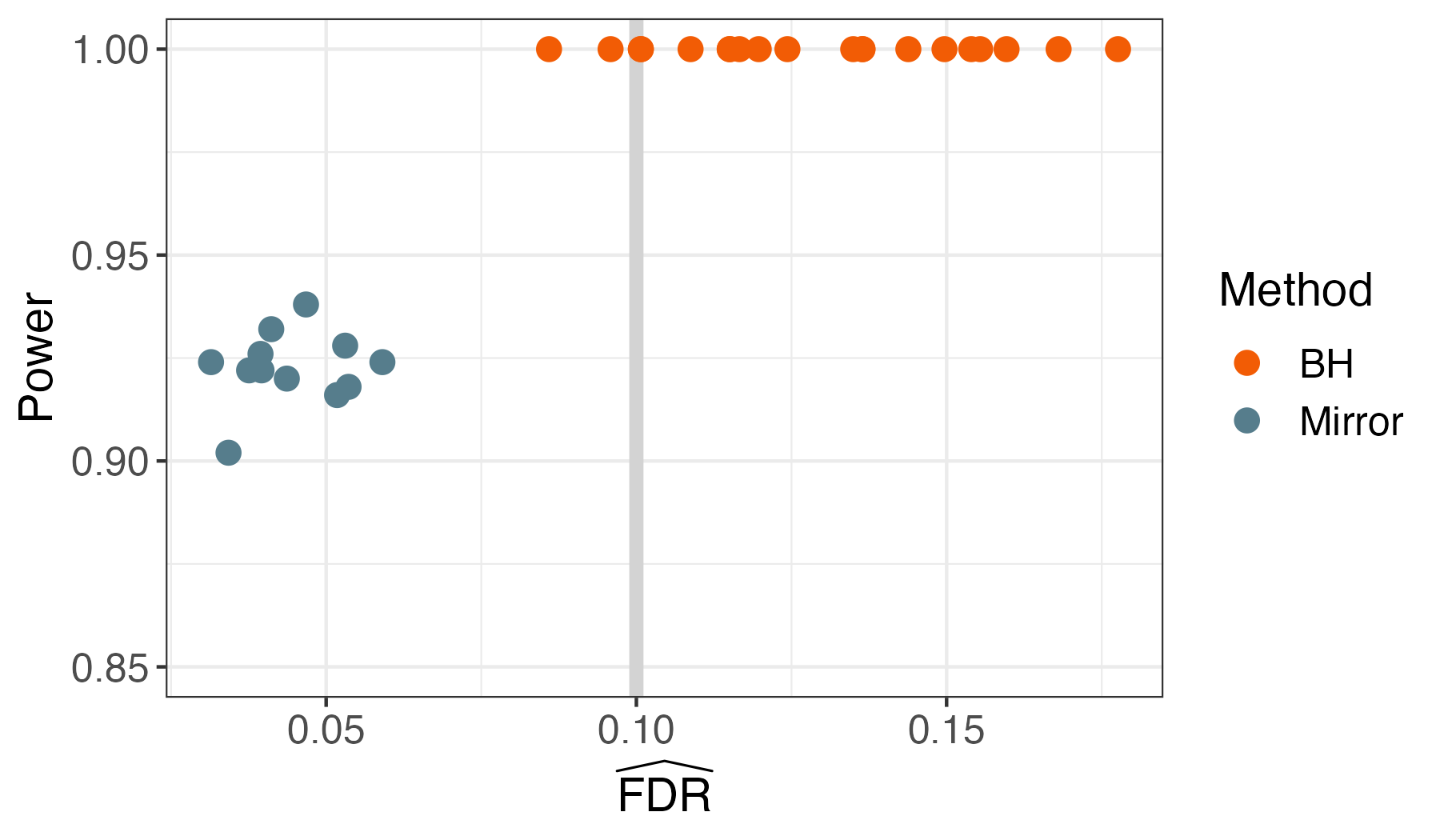
Teaser: Microbiome Intervention Analysis
Mirror statistics support formal inference of delayed, nonlinear intervention effects in longitudinal microbiome studies [12].

Manuscript, Documentation, Demo
Conclusion
- Whenever the signal is subtle, hypothesis testing is crucial.
- The key abstractions we encountered were,
- Optimization view of BH + Context = Focused Hypothesis Testing
- Data splitting + Symmetry assumptions = Formal Error Control
- These abstractions are useful across the computational genomics workflow
- E.g., in experimental design [13] and interpretation of features generated through unsupervised learning [14; 11].
- Other variations developed in my lab [12; 15; 16]
References
[1] Y. Benjamini and Y. Hochberg. "Controlling the false discovery rate: a practical and powerful approach to multiple testing". In: Journal of the royal statistical society series b-methodological 57 (1995), pp. 289-300.
[2] N. Ignatiadis, B. Klaus, J. B. Zaugg, et al. "Data-driven hypothesis weighting increases detection power in genome-scale multiple testing". In: Nature methods 13 (2016), pp. 577 - 580.
[3] L. Lei and W. Fithian. "AdaPT: an interactive procedure for multiple testing with side information". In: Journal of the Royal Statistical Society: Series B (Statistical Methodology) 80 (2016).
[4] N. Ignatiadis and W. Huber. "Covariate powered cross‐weighted multiple testing". In: Journal of the Royal Statistical Society: Series B (Statistical Methodology) 83 (2017).
References
[5] M. J. Zhang, F. Xia, and J. Y. Zou. "Fast and covariate-adaptive method amplifies detection power in large-scale multiple hypothesis testing". In: Nature Communications 10 (2019).
[6] R. Yurko, M. G. G'Sell, K. Roeder, et al. "A selective inference approach for false discovery rate control using multiomics covariates yields insights into disease risk". In: Proceedings of the National Academy of Sciences of the United States of America 117 (2020), pp. 15028 - 15035.
[7] M. D. Robinson, D. J. McCarthy, and G. K. Smyth. "edgeR: a Bioconductor package for differential expression analysis of digital gene expression data". In: Bioinformatics 26 (2009), pp. 139 - 140.
[8] M. Stephens. "Simple Transform Simulation". In: Online (2023). Accessed: 2023-07-14.
References
[9] C. Dai, B. Lin, X. Xing, et al. "False Discovery Rate Control via Data Splitting". In: Journal of the American Statistical Association (2020).
[10] X. Ge, Y. E. Chen, D. Song, et al. "Clipper: p-value-free FDR control on high-throughput data from two conditions". In: Genome Biology 22 (2021).
[11] F. R. Guo and R. D. Shah. "Rank-transformed subsampling: inference for multiple data splitting and exchangeable p-values". In: ArXiv (2023).
[12] K. Sankaran and P. Jeganathan. "Microbiome Intervention Analysis with Transfer Functions and Mirror Statistics". In: ArXiv (2023).
References
[13] C. Fannjiang, S. Bates, A. N. Angelopoulos, et al. "Conformal prediction under feedback covariate shift for biomolecular design". In: Proceedings of the National Academy of Sciences of the United States of America 119 (2022).
[14] D. Song and J. J. Li. "PseudotimeDE: inference of differential gene expression along cell pseudotime with well-calibrated p-values from single-cell RNA sequencing data". In: Genome Biology 22 (2021).
[15] K. Sankaran and S. P. Holmes. "structSSI: Simultaneous and Selective Inference for Grouped or Hierarchically Structured Data." In: Journal of statistical software 59 13 (2014), pp. 1-21 .
[16] K. Sankaran. "Functions for Performing Local FDR Estimation when Null and Alternative are Poisson [R package LocFDRPois version 1.0.0]". In: CRAN (2015).
Selective Inference
Step (1) distinguishes selective inference from ordinary statistical inference.
- Search for interesting patterns.
- Test whether they could have just been coincidences.
Looking for promising contexts \implies selective inference.
Optimization
This can be given into generic optimization solvers:
\begin{align*} \text{maximize } & \sum_{g} m_{g}\hat{F}\left(t_{g}\right) \\ \text{subject to } & \sum_{g = 1}^{G} m_{g}\left(t_{g} - m_{g}\hat{F}\left(t_{g}\right)\right) \leq q \end{align*}
Aggregation Procedure
We need a mechanism for aggregating \hat{S}_{k} across iterations.
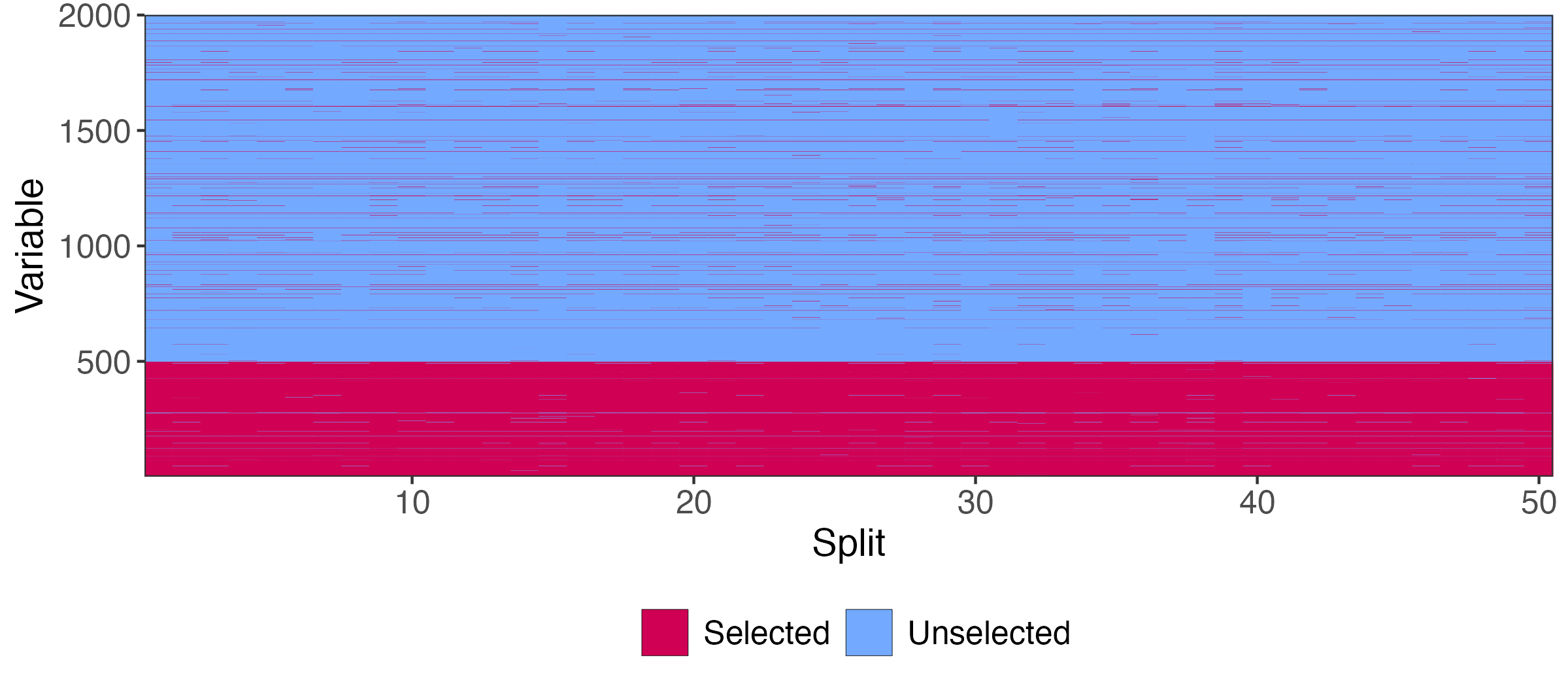
Aggregation Procedure
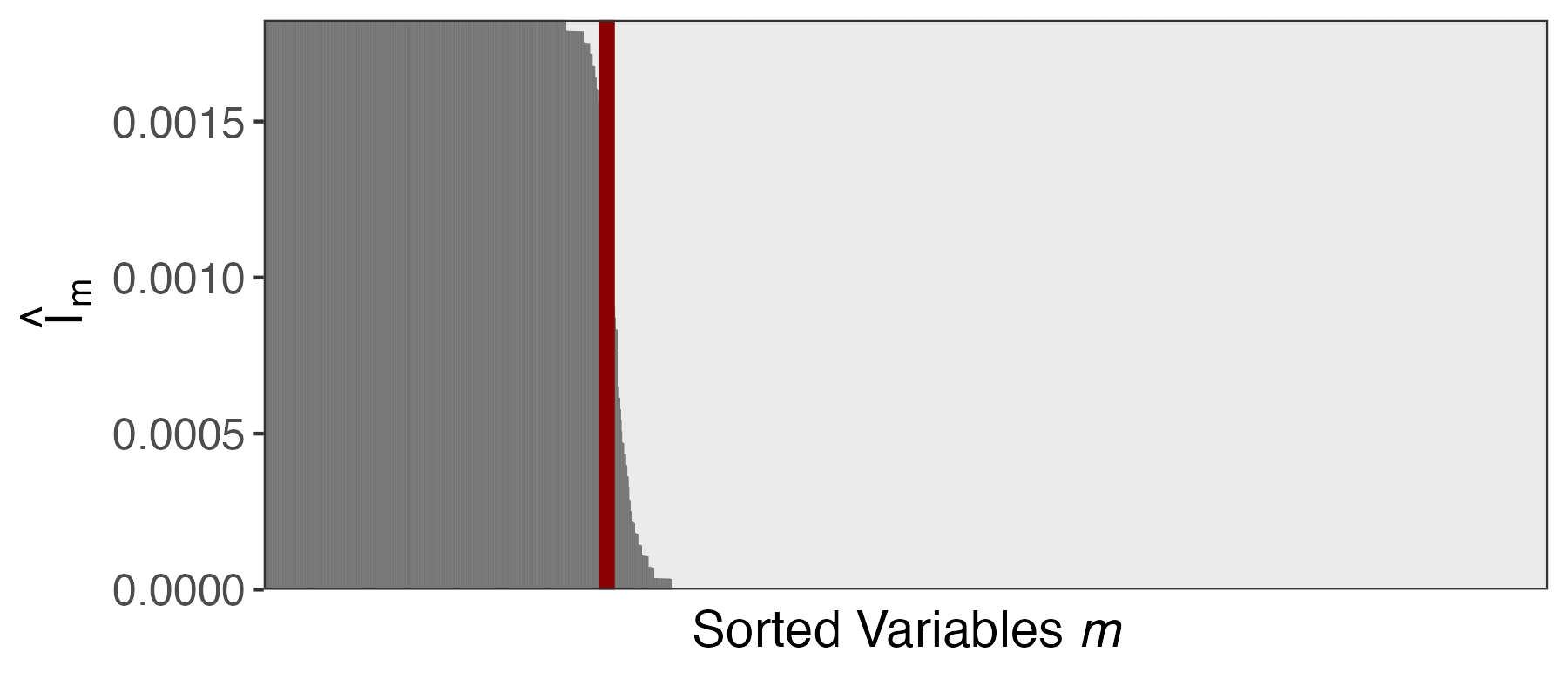
Let \hat{I}_{1}, \dots, \hat{I}_{m} store the row averages of this matrix. Larger \hat{I}_{m} means:
- The feature is selected even when \absarg{\hat{S}_{k}} is small
- The feature is selected frequently

Aggregation Procedure
Let \hat{I}_{1}, \dots, \hat{I}_{m} store the column averages of this matrix. Larger \hat{I}_{m} means:
- The feature is selected even when \absarg{\hat{S}_{k}} is small
- The feature is selected frequently
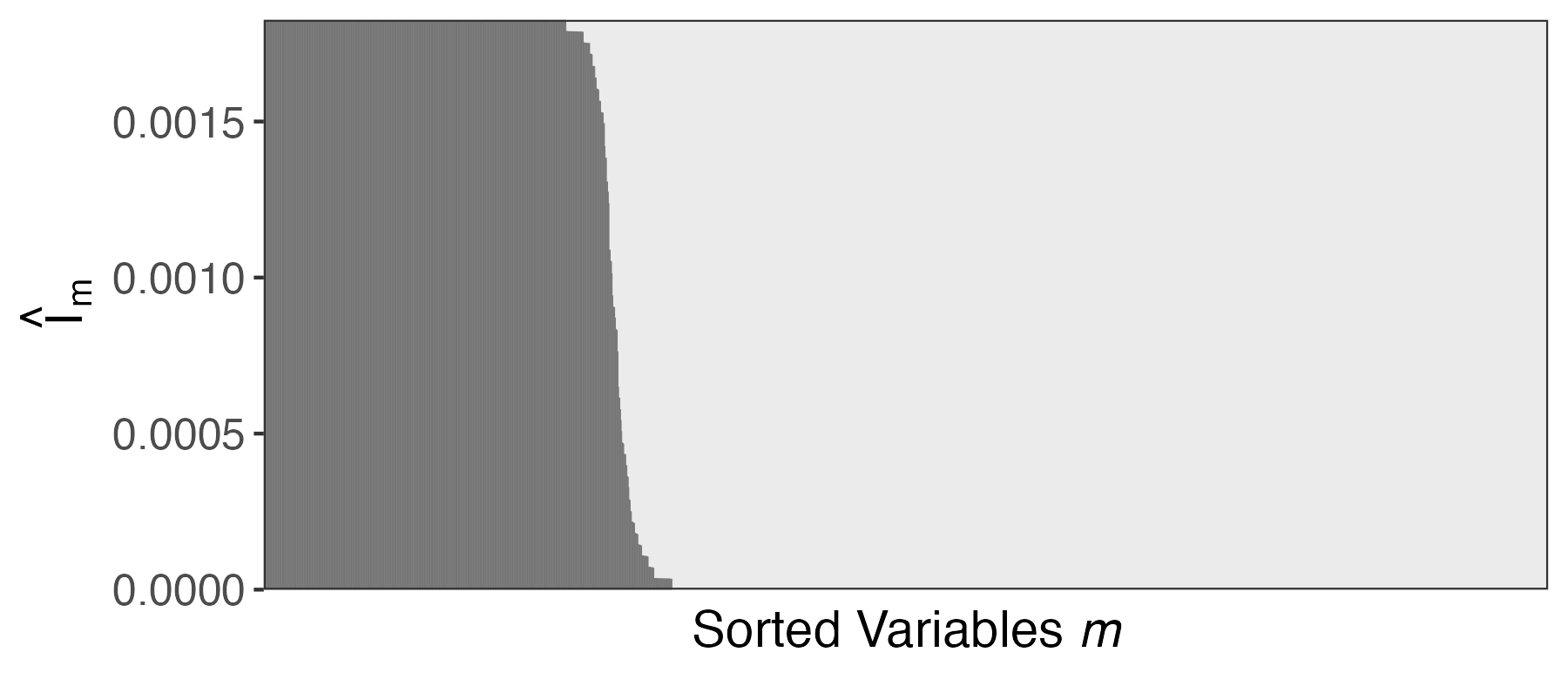
Aggregation Procedure
- For the final selection, choose features with the largest \hat{I}_{m} possible, up until the budget exceeds 1 - q.
- This is guaranteed to (asymptotically) control the FDR at level q